- TCGA Cancer Type Abbreviations
- miRNA Clusters
- CmirC
- Workflow of Data Browsing Modules
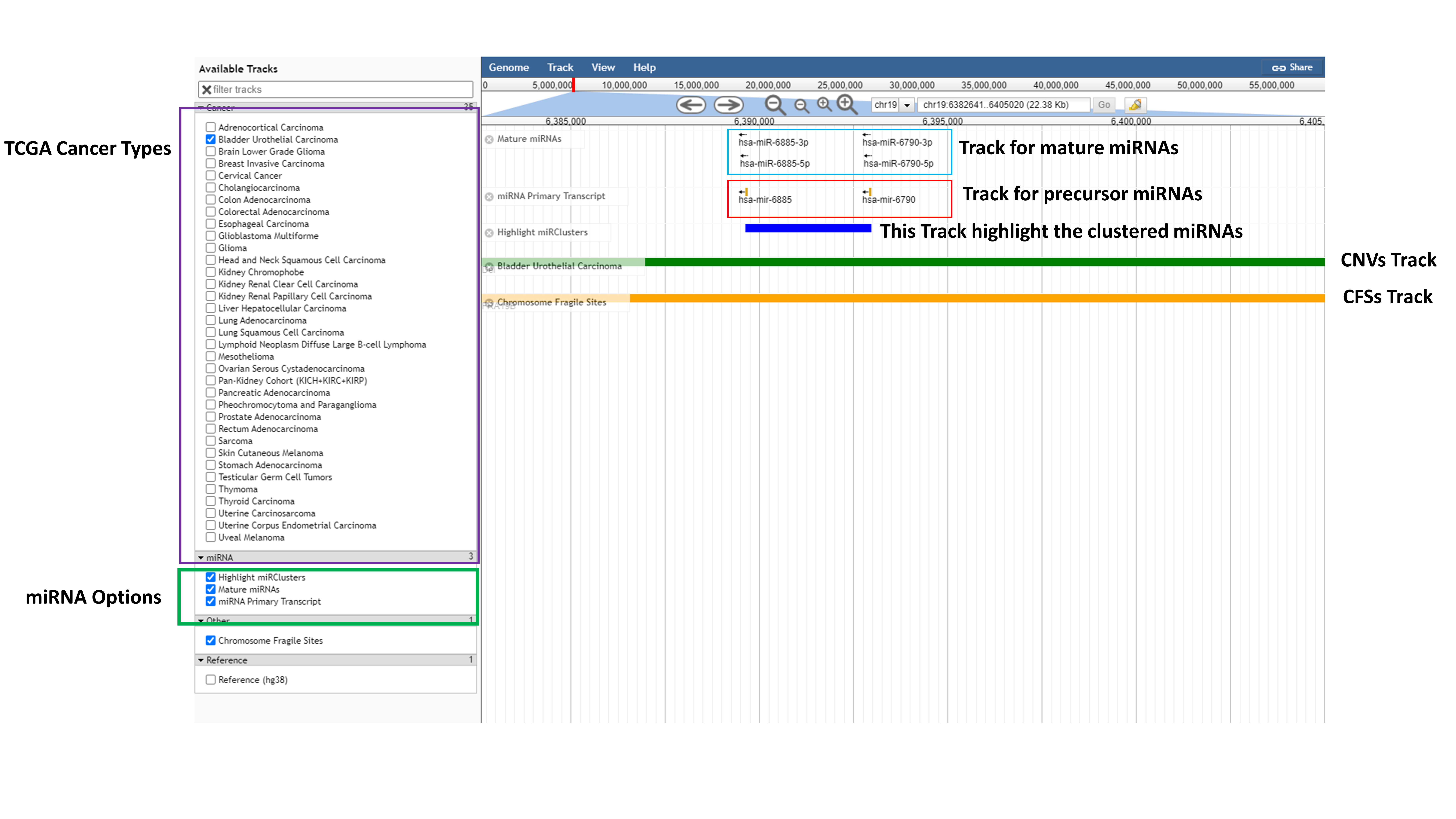
- Genome Browser
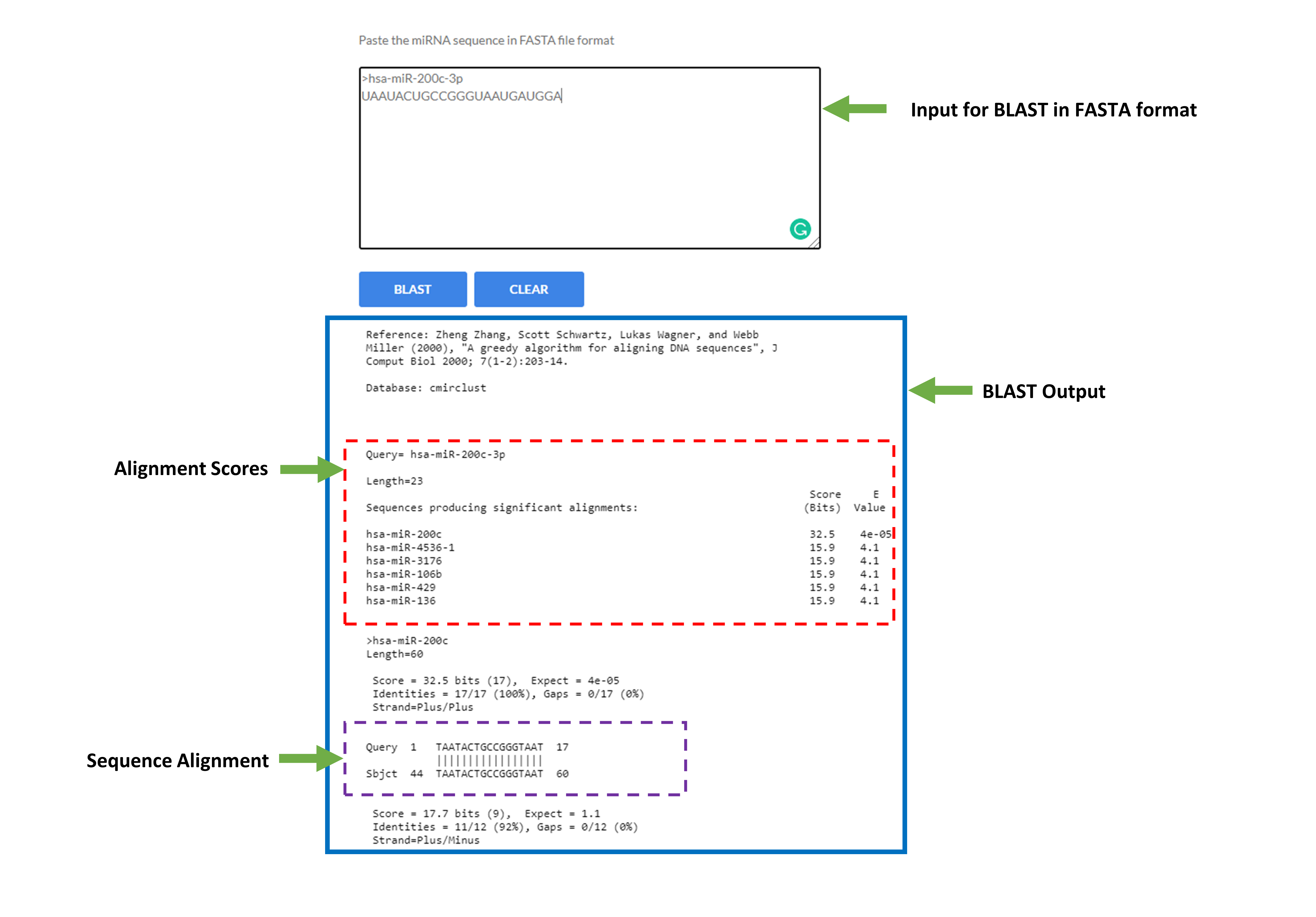
- Basic Local Alignment Search Tool (BLAST)
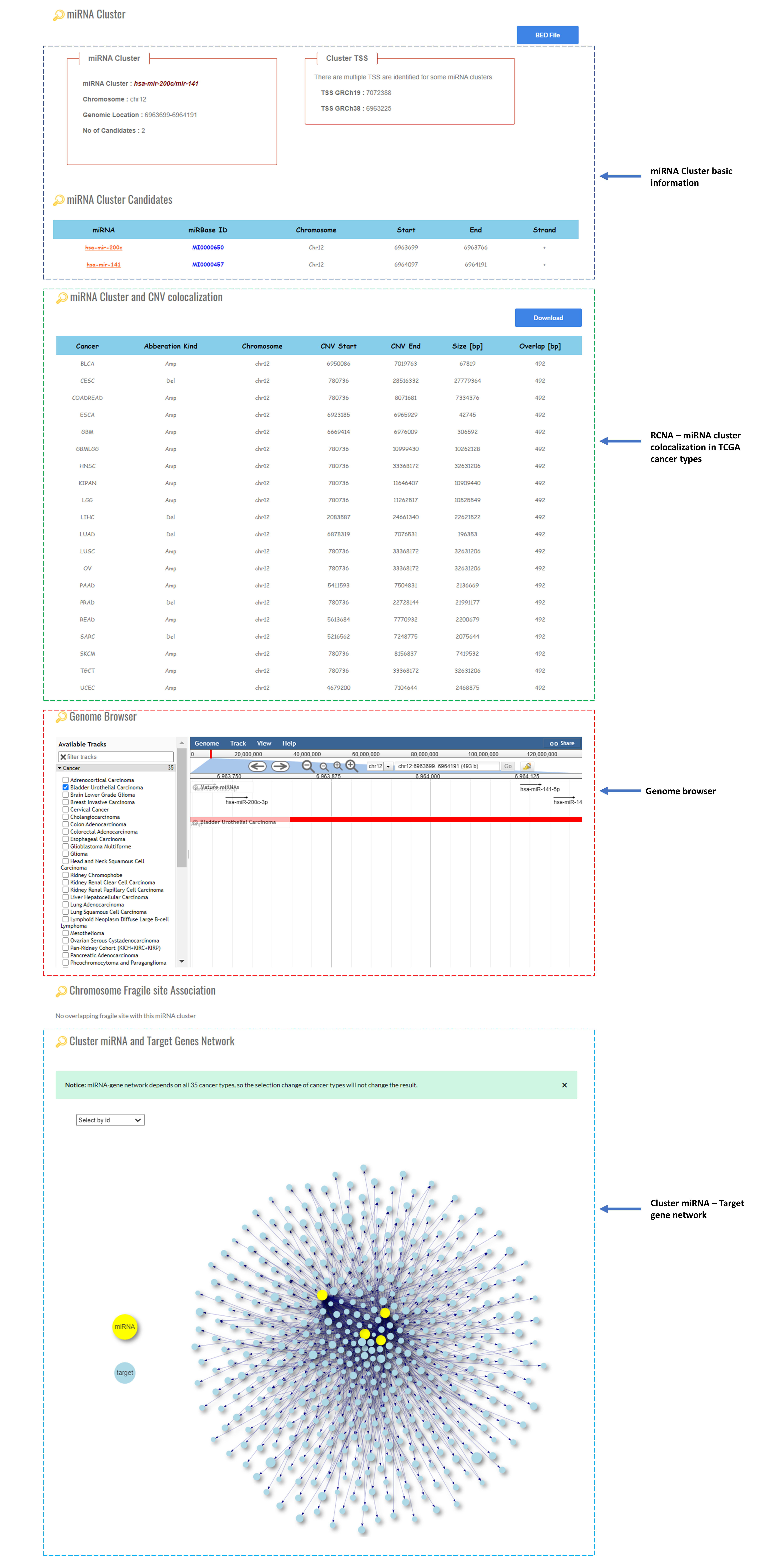
- miRNA Cluster Details Page
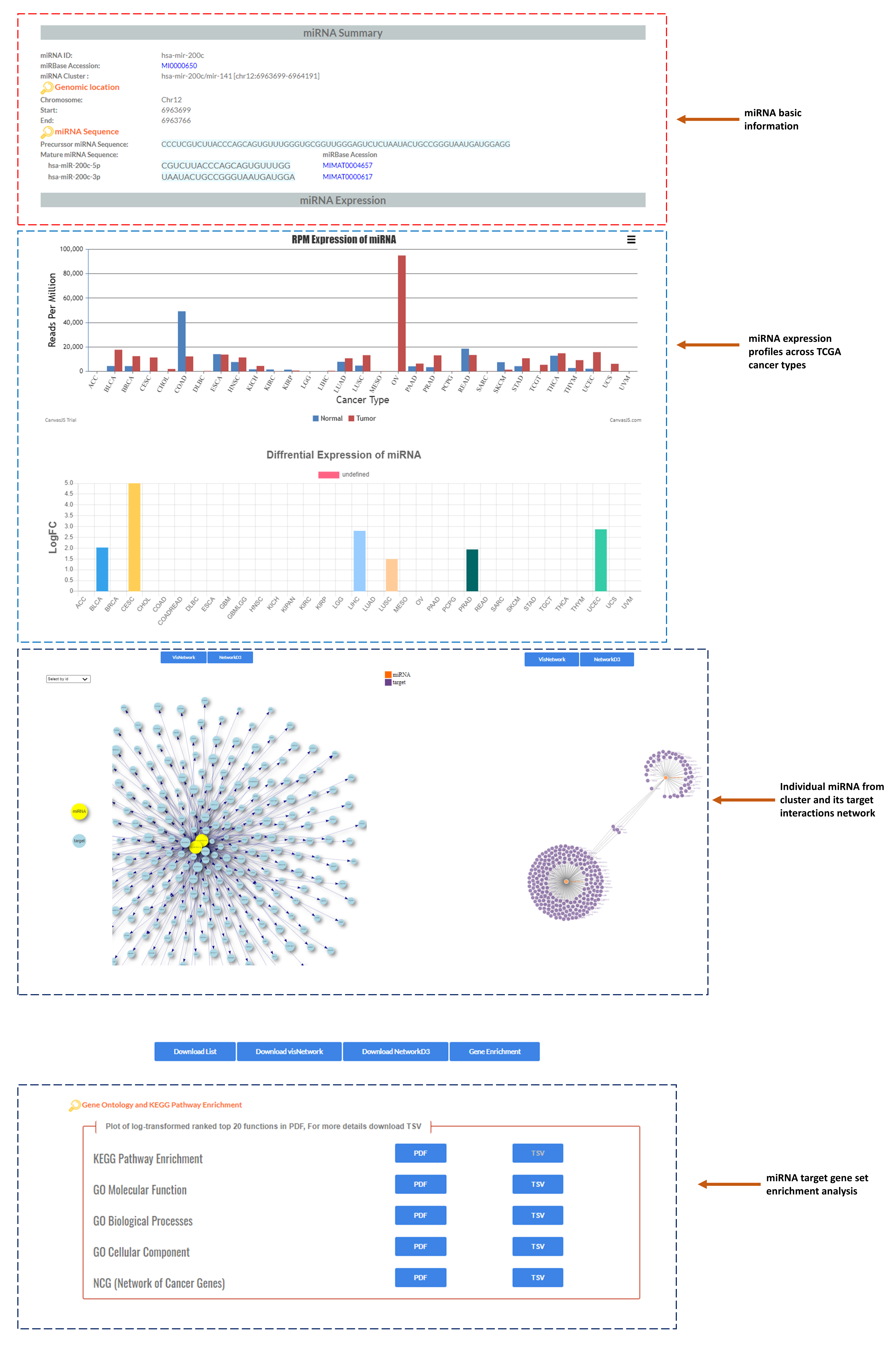
- Individual miRNA Details Page
| Sr. No. | Abbreviation | Cancer Type |
|---|---|---|
| 1 | TCGA-ACC | Adrenocortical carcinoma |
| 2 | TCGA-BLCA | Bladder urothelial carcinoma |
| 3 | TCGA-BRCA | Breast invasive carcinoma |
| 4 | TCGA-CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma |
| 5 | TCGA-CHOL | Cholangiocarcinoma |
| 6 | TCGA-COAD | Colon adenocarcinoma |
| 7 | TCGA-COADREAD | Colorectal adenocarcinoma |
| 8 | TCGA-DLBC | Lymphoid Neoplasm Diffuse Large B-cell Lymphoma |
| 9 | TCGA-ESCA | Esophageal carcinoma |
| 10 | TCGA-GBM | Glioblastoma multiforme |
| 11 | TCGA-GBMLGG | Glioma |
| 12 | TCGA-HNSC | Head and Neck squamous cell carcinoma |
| 13 | TCGA-KICH | Kidney Chromophobe |
| 14 | TCGA-KIPAN | Pan-kidney cohort(KICH+KIRC+KIRP) |
| 15 | TCGA-KIRC | Kidney renal clear cell carcinoma |
| 16 | TCGA-KIRP | Kidney renal papillary cell carcinoma |
| 17 | TCGA-LGG | Brain Lower Grade Glioma |
| 18 | TCGA-LIHC | Liver hepatocellular carcinoma |
| 19 | TCGA-LUAD | Lung adenocarcinoma |
| 20 | TCGA-LUSC | Lung squamous cell carcinoma |
| 21 | TCGA-MESO | Mesothelioma |
| 22 | TCGA-OV | Ovarian serous cystadenocarcinoma |
| 23 | TCGA-PAAD | Pancreatic adenocarcinoma |
| 24 | TCGA-PCPG | Pheochromocytoma and Paraganglioma |
| 25 | TCGA-PRAD | Prostate adenocarcinoma |
| 26 | TCGA-READ | Rectum adenocarcinoma |
| 27 | TCGA-SARC | Sarcoma |
| 28 | TCGA-SKCM | Skin Cutaneous Melanoma |
| 29 | TCGA-STAD | Stomach adenocarcinoma |
| 30 | TCGA-TGCT | Testicular Germ Cell Tumors |
| 31 | TCGA-THCA | Thyroid carcinoma |
| 32 | TCGA-THYM | Thymoma |
| 33 | TCGA-UCEC | Uterine Corpus Endometrial Carcinoma |
| 34 | TCGA-UCS | Uterine Carcinosarcoma |
| 35 | TCGA-UVM | Uveal Melanoma |
Generally, a miRNA cluster consists of two or more miRNAs residing neighborly on the same strand. miRNAs in a cluster are not separated by a transcription unit and transcribed in the same orientation. The miRNAs reported from the clusters are co-expressed and jointly regulate functionally associated genes. Some studies have reported that clustered miRNAs are transcribed as poly-cistrons and have similar expression patterns.The intergenomic distance between two miRNAs is considered to group miRNAs as a cluster. According to the miRbase repository, more than 40% of experimentally validated human miRNA cluster genes have been identified within the 10 kb region.
CmirC is a cancer omics database which provides information on recurrent copy number variation (RCNV) co-localized miRNA clusters from 35 TCGA cancer types. Here we are presenting the primary version of CmirC which will be further expanded according to subsequent TCGA data update
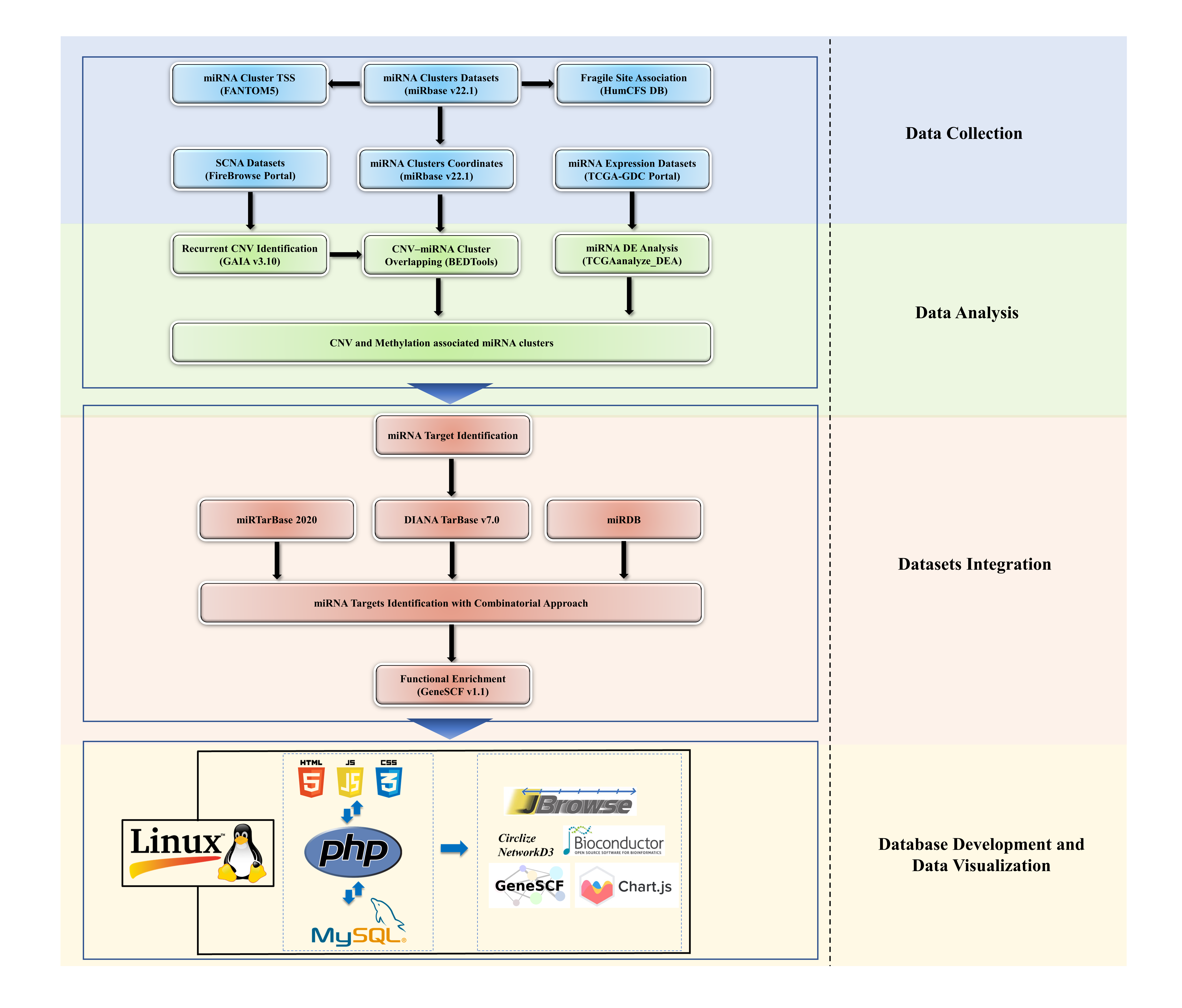
Data browsing modules allows user to extensively browse data stored in database with multiple options:
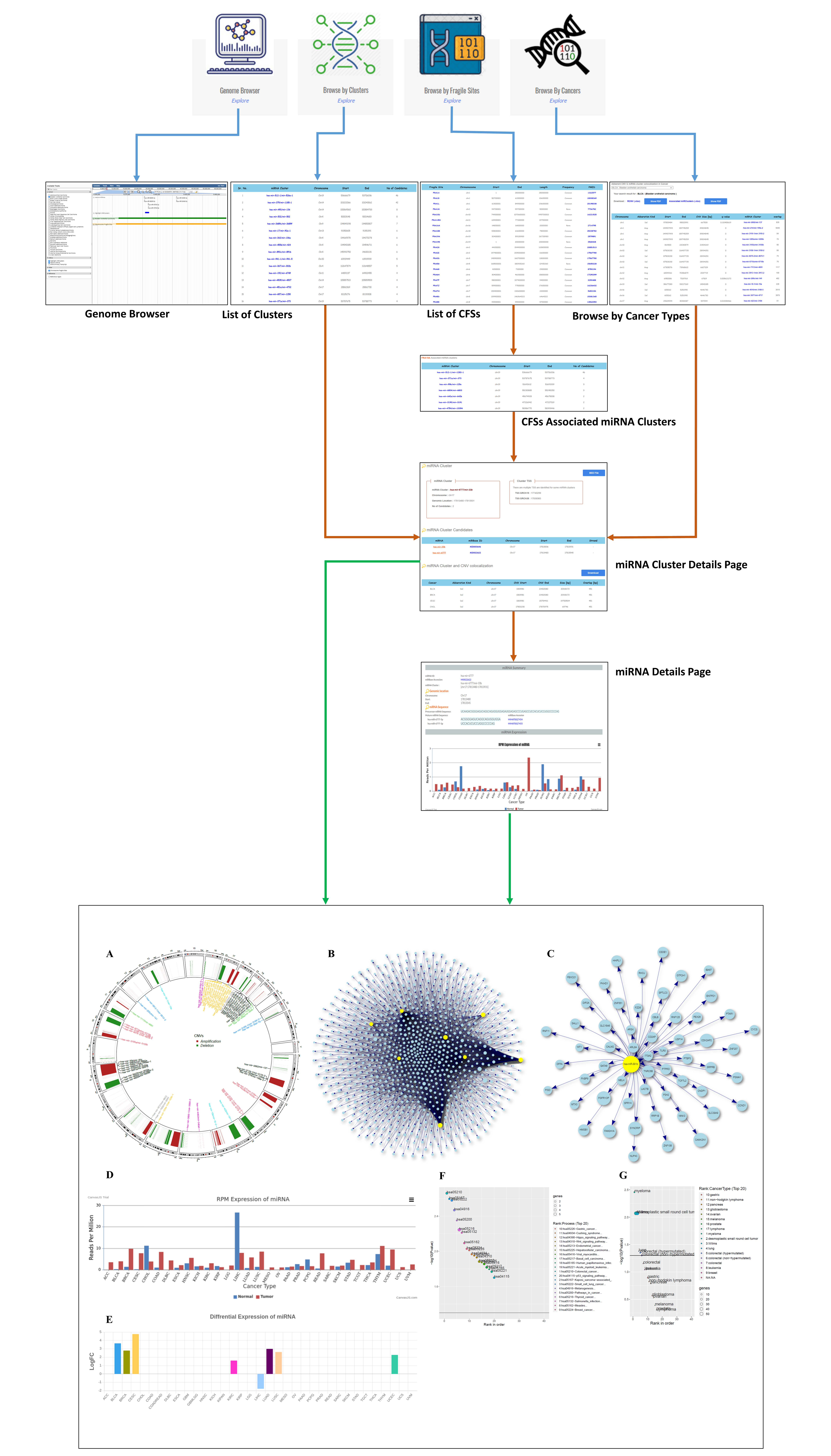
Interactive data visualization by CmirC. A. Circos plot illustrating identified RCNA and co-localized miRNA Clusters B. miRNA Cluster and target gene network C. Individual miRNA targets D. Average read per million (RPM) expression of miRNA across TCGA cancer types E. miRNA differential expression (Normal vs Tumor) across the TCGA cancer types F. Top 20 enriched pathways for gene set targeted by miRNA clusters G. Gene set enriched in the cancer network
Genome browser module is developed to visualize the RCNA and miRNA cluster co-localization across 35 TCGA cancer types. Here user can select particular TCGA cancer types, Chromosome Fragile Sites, mature or precursor miRNAs and can highlight the clustered miRNAs. By clicking on zoom function user can also see the miRNA name and by clicking on miRNA, on pop up window user can see the information available in database for that particular miRNA.
BLAST module is developed for performing BLAST search against nucleotide sequence of Clustered miRNAs. Users are requested to submit sequence only in FASTA format. This is a nucleotide to nucleotide search, so BLASTN will be used to perform search.
 rC
rC