An integrated pipeline for identification of clustered miRNAs co-localized with RCNA
How it works?
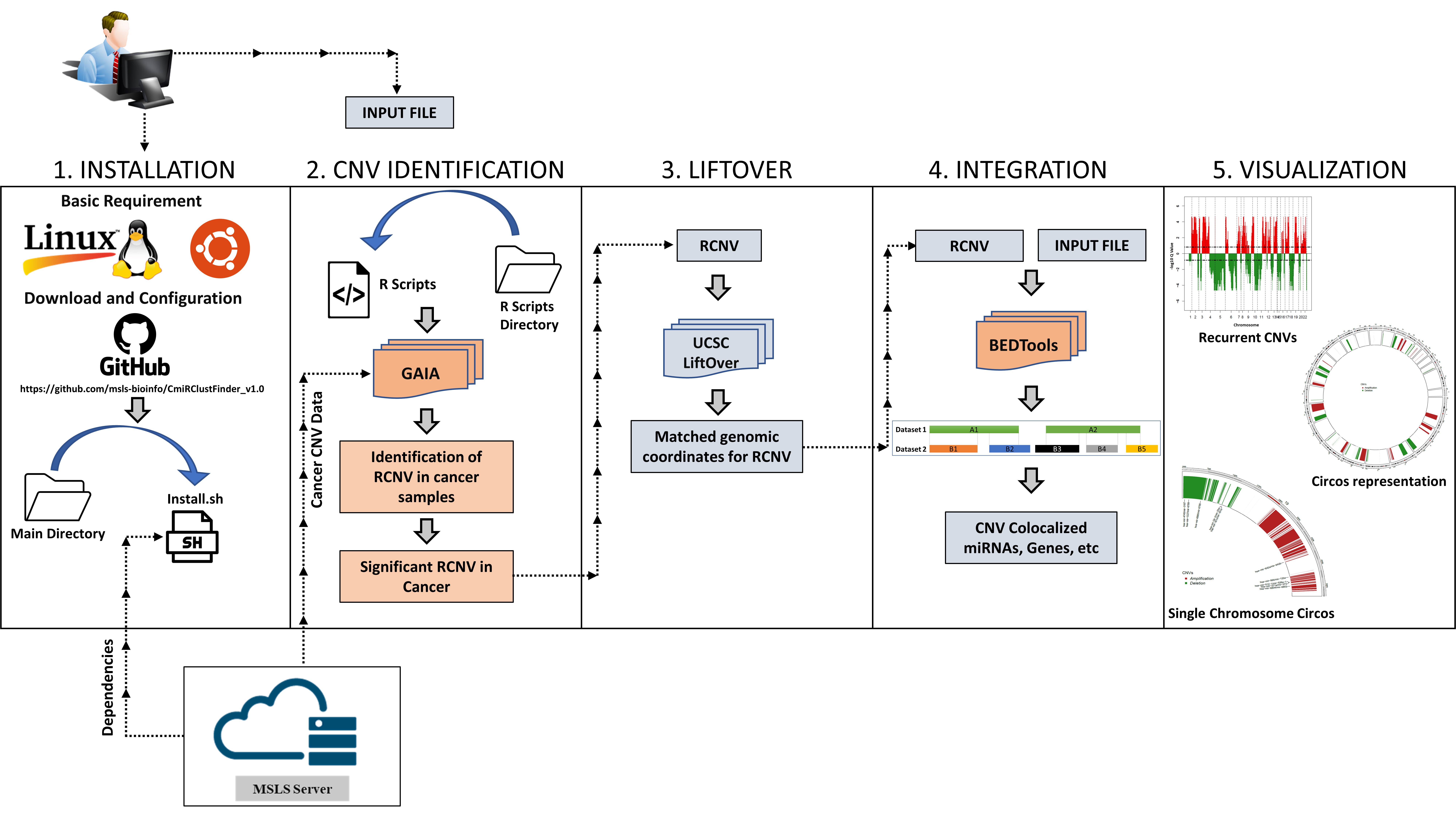
CmiRClustFinder v1.0 computes the integrated data in five steps. The installation script will download all the necessary resources and prepare the pipeline for use in the first step. In the second step, the GAIA package finds frequent aberrations in chromosomal regions among cancer patients’ datasets. In the third step, the LiftOver tool matches the genomic build for RCNVs and user-defined genetic elements. We have integrated BEDTools to find co-localization of significant RCNV and genomic elements in the fourth step. Lastly, the Circlize package generates a circos representation of the data.